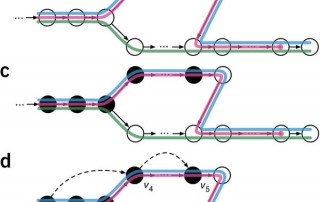
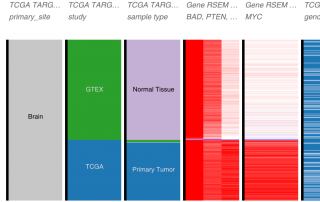
A word about unwanted variability
Experiments are influenced by various variables: the one we are interested in, and many others. Variability in the data can be related to differences in technical or biological variables, such as the instrument used, genetic background, age, gender, etc. Therefore, batch effects are frequently observed in gene expression datasets. They can affect genes independently of the variable of interest (independent of cancer or normal states, for example) or not (the expression of a given gene might be influenced by the [...]