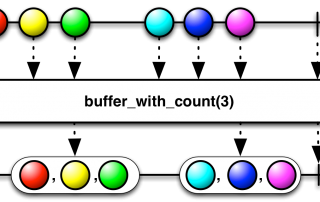
Document your work by adding parameters to your shell scripts
At some point during your bioinformatics career, you're going to start writing shell scripts, it's kind of inevitable ! So let us discuss a strategy to add parameters to your scripts in order make them more easily reusable, while also keeping a trace of the settings you used to generate a set of results. (Disclaimer: the procedures described below have been tested using BASH, some modifications might be necessary if you use a different shell.) Modifying your scripts in order [...]